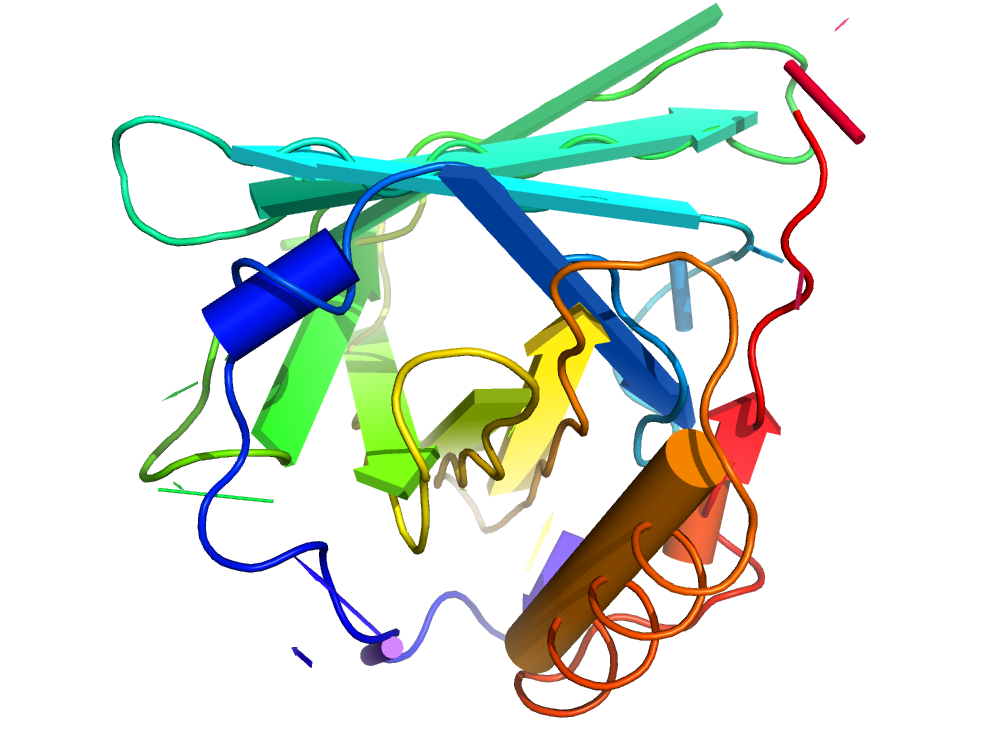
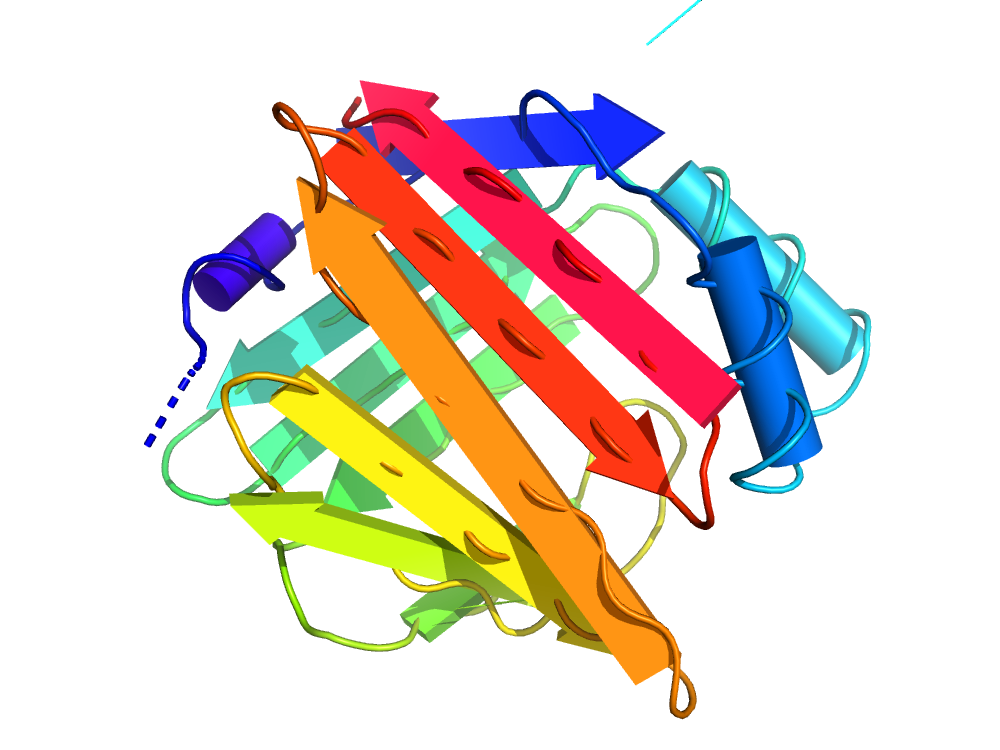
OverProt creates an overview of secondary structure elements in protein families.
For each protein family from the CATH database, a secondary structure consensus is available, showing the characteristic helices and β-sheets of the family. A consensus for an arbitrary subset of a family can be computed using User-defined queries.
Search
Search the precomputed OverProt results for the structural families in the CATH database.
Enter a CATH family ID (e.g. 1.10.630.10) or PDB ID (e.g. 2nnj) or CATH domain ID (e.g. 2nnjA00).
User-defined queries
Run OverProt on your own list of protein structures ⇒
Examples
Cytochrome P450
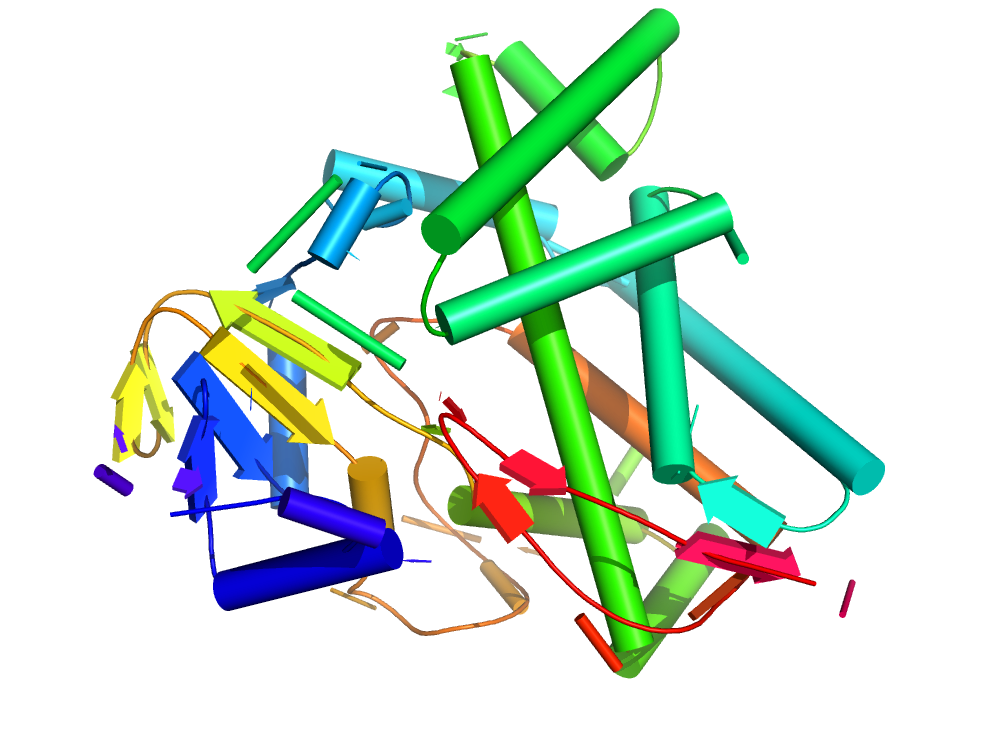
Cytochromes P450 (CYPs) are a family of enzymes (CATH ID 1.10.630.10) containing heme as a cofactor that function as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important e.g. for the clearance of various compounds. In plants, they are involved in the biosynthesis of defensive compounds, fatty acids, and hormones.
Dipeptidyl peptidase IV
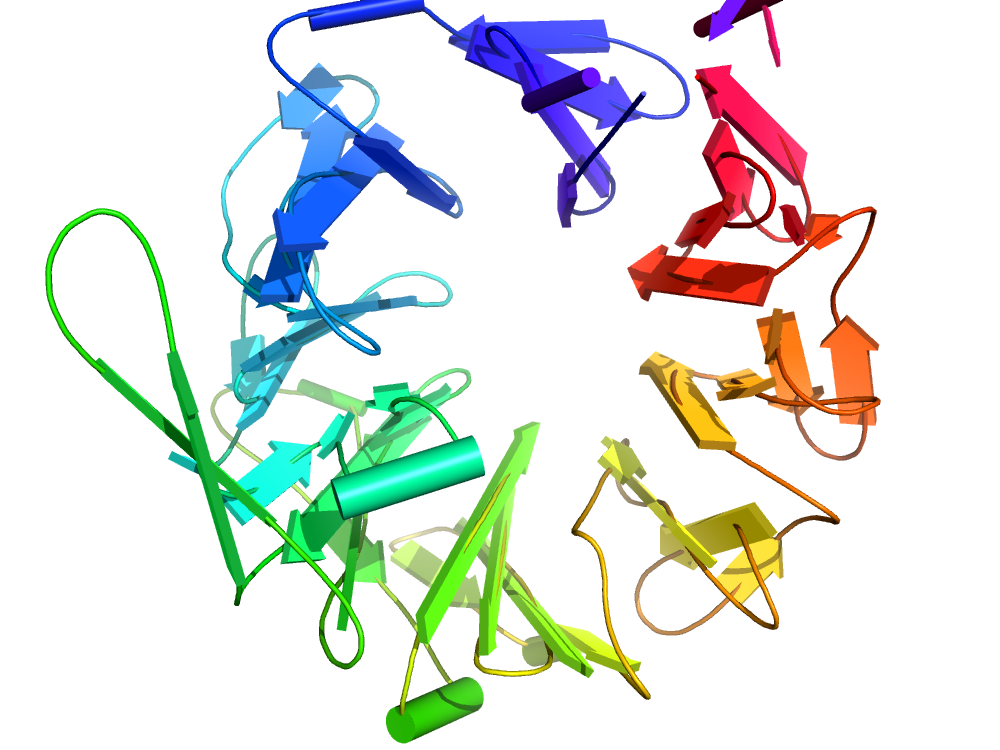
Dipeptidyl peptidase IV is a serine protease, it contributes to the regulation of various physiological processes and serves as a marker of autoimmune diseases, adenosine deaminase-deficiency and HIV. This example shows its eight-bladed β-propeller domain (CATH ID 2.140.10.30), important for a function of the enzyme.
Lipocalin
Lipocalins are extracellular proteins which function include transport of nutrient or pheromones, control of cell regulation, etc. The most common structure of the lipocalin protein fold is a single eight-stranded antiparallel beta-sheet, forming a continuously hydrogen-bonded beta-barrel (see left figure). But also a ten-stranded fold occurs frequently (see right figure). OverProt user-defined queries enable to analyse representatives (from CATH Superfamily 2.40.128.20) of both these types separately and compute a consensus sequence for them. Here we provide input queries for eight-stranded and ten-stranded lipocalins.
API documentation
Overview of the available machine-readable data ⇒
Citing
If you found SecStrAnnotator helpful, please cite:
- Midlik A, Hutařová Vařeková I, Hutař J, Chareshneu A, Berka K, Svobodová R (2022) OverProt: secondary structure consensus for protein families. Bioinformatics, 38, 3648–3650. https://doi.org/10.1093/bioinformatics/btac384
Contact
If you have any feedback to the algorithm, visualization, or the results for a specific family, please contact us:
midlik@mail.muni.cz, Laboratory of Computational Chemistry, Masaryk University
License
All data provided by this site are freely available to all users. The underlying software is open source and available at GitLab under MIT license.